Machine learning is not as abstract as one might think. If you want to get value out of known data and do predictions for unknown data, the most important challenge is asking the right questions and of course knowing what you are doing, especially if you want to optimize your prediction accuracy.
In this blog I'm exploring an example of machine learning. The random forest algorithm. I'll provide an example on how you can use this algorithm to do predictions. In order to implement a random forest, I'm using R with the randomForest library and I'm using the iris dataset which is provided by the R installation.
The Random Forest
A popular method of machine learning is by using decision tree learning. Decision tree learning comes closest to serving as an off-the-shelf procedure for data mining (see here). You do not need to know much about your data in order to be able to apply this method. The random forest algorithm is an example of a decision tree learning algorithm.
Random forest in (very) short
How it works exactly takes some time to figure out. If you want to know details, I recommend watching some youtube recordings of lectures on the topic. Some of its most important features of this method:
- A random forest is a method to do classifications based on features. This implies you need to have features and classifications.
- A random forest generates a set of classification trees (an ensemble) based on splitting a subset of features at locations which maximize information gain. This method is thus very suitable for distributed parallel computation.
- Information gain can be determined by how accurate the splitting point is in determining the classification. Data is split based on the feature at a specific point and the classification on the left and right of the splitting point are checked. If for example the splitting point splits all data of a first classification from all data of a second classification, the confidence is 100%; maximum information gain.
- A splitting point is a branching in the decision tree.
- Splitting points are based on values of features (this is fast)
- A random forest uses randomness to determine features to look at and randomness in the data used to construct the tree. Randomness helps reducing compute time.
- Each tree gets to see a different dataset. This is called bagging.
- Tree classification confidences are summed and averaged. Products of the confidences can also be taken. Individual trees have a high variance because they have only seen a small subset of data. Averaging helps creating a better result.
- With correlated features, strong features can end up with low scores and the method can be biased towards variables with many categories.
- A random forest does not perform well with unbalanced datasets; samples where there are more occurrences of a specific class.
Use cases for a random forest can be for example text classification such as spam detection. Determine if certain words are present in a text can be used as a feature and the classification would be spam/not spam or even more specific such as news, personal, etc. Another interesting use case lies in genetics. Determining if the expression of certain genes is relevant for a specific disease. This way you can take someone's DNA and determine with a certain confidence if someone will contract a disease. Of course you can also take other features into account such as income, education level, smoking, age, etc.
R
Why R
I decided to start with R. Why? Mainly because it is easy. There are many libraries available and there is a lot of experience present worldwide; a lot of information can be found online. R however also has some drawbacks.
Some benefits
- It is free and easy to get started. Hard to master though.
- A lot of libraries are available. R package management works well.
- R has a lot of users. There is a lot of information available online
- R is powerful in that if you know what you are doing, you require little code doing it.
Some challenges
- R loads datasets in memory
- R is not the best at doing distributed computing but can do so. See for example here
- The R syntax can be a challenge to learn
To get a server to play with, I decided to go with Ubuntu Server. I first installed the usual things like a GUI. Next I installed some handy things like a terminal emulator, firefox and stuff like that. I finished with installing R and R-studio; the R IDE.
So first download and install Ubuntu Server (next, next, finish)
sudo apt-get update
sudo apt-get install aptitude
--Install a GUI
sudo aptitude install --without-recommends ubuntu-desktop
-- Install the VirtualBox Guest additions
sudo apt-get install build-essential linux-headers-$(uname -r)
Install guest additions (first mount the ISO image which is part of VirtualBox, next run the installer)
-- Install the below stuff to make Dash (Unity search) working
http://askubuntu.com/questions/125843/dash-search-gives-no-result
sudo apt-get install unity-lens-applications unity-lens-files
-- A shutdown button might come in handy
sudo apt-get install indicator-session
-- Might come in handy. Browser and fancy terminal application
sudo apt-get install firefox terminator
--For the installation of R I used the following as inspiration: https://www.r-bloggers.com/how-to-install-r-on-linux-ubuntu-16-04-xenial-xerus/
sudo echo "deb http://cran.rstudio.com/bin/linux/ubuntu xenial/" | sudo tee -a /etc/apt/sources.list
gpg --keyserver keyserver.ubuntu.com --recv-key E084DAB9
gpg -a --export E084DAB9 | sudo apt-key add -
sudo apt-get update
sudo apt-get install r-base r-base-dev
-- For the installation of R-studio I used: https://mikewilliamson.wordpress.com/2016/11/14/installing-r-studio-on-ubuntu-16-10/
wget http://ftp.ca.debian.org/debian/pool/main/g/gstreamer0.10/libgstreamer0.10-0_0.10.36-1.5_amd64.deb
wget http://ftp.ca.debian.org/debian/pool/main/g/gst-plugins-base0.10/libgstreamer-plugins-base0.10-0_0.10.36-2_amd64.deb
sudo dpkg -i libgstreamer0.10-0_0.10.36-1.5_amd64.deb
sudo dpkg -i libgstreamer-plugins-base0.10-0_0.10.36-2_amd64.deb
sudo apt-mark hold libgstreamer-plugins-base0.10-0
sudo apt-mark hold libgstreamer0.10
wget https://download1.rstudio.org/rstudio-1.0.136-amd64.deb
sudo dpkg -i rstudio-1.0.136-amd64.deb
sudo apt-get -f install
Doing a random forest in R
R needs some libraries to do random forests and create nice plots. First give the following commands:
#to do random forests
install.packages("randomForest")
#to work with R markdown language
install.packages("knitr")
#to create nice plots
install.packages("ggplot2")
In order to get help on a library you can give the following command which will give you more information on the library.
library(help = "randomForest")
Of course, the randomForest implementation does have some specifics:
- it uses the reference implementation based on CART trees
- it is biased in favor of continuous variables and variables with many categories
A simple program to do a random forest looks like this:
#load libraries
library(randomForest)
library(knitr)
library(ggplot2)
#random numbers after the set.seed(10) are reproducible if I do set.seed(10) again
set.seed(10)
#create a training sample of 45 items from the iris dataset. replace indicates items can only be present once in the dataset. If replace is set to true, you will get Out of bag errors.
idx_train <- sample(1:nrow(iris), 45, replace = FALSE)
#create a data.frame from the data which is not in the training sample
tf_test <- !1:nrow(iris) %in% idx_train
#the column ncol(iris) is the last column of the iris dataset. this is not a feature column but a classification column
feature_columns <- 1:(ncol(iris)-1)
#generate a randomForest.
#use the feature columns from training set for this
#iris[idx_train, ncol(iris)] indicates the classification column
#importance=TRUE indicates the importance of features in determining the classification should be determined
#y = iris[idx_train, ncol(iris)] gives the classifications for the provided data
#ntree=1000 indicates 1000 random trees will be generated
model <- randomForest(iris[idx_train, feature_columns], y = iris[idx_train, ncol(iris)], importance = TRUE, ntree = 1000)
#print the model
#printing the model indicates how the sample dataset is distributed among classes. The sum of the sample classifications is 45 which is the sample size. OOB rate indicates 'out of bag' (the overall classification error).
print(model)
#we use the model to predict the class based on the feature columns of the dataset (minus the sample used to train the model).
response <- predict(model, iris[tf_test, feature_columns])
#determine the number of correct classifications
correct <- response == iris[tf_test, ncol(iris)]
#determine the percentage of correct classifications
sum(correct) / length(correct)
#print a variable importance (varImp) plot of the randomForest
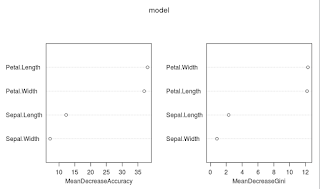
varImpPlot(model)
#in this dataset the petal length and width are more important measures to determine the class than the sepal length and width.



No comments:
Post a Comment